Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Theoretical Investigation on Rh(III)- And Pd(II)- Catalyzed Regioselective Oxidative Annulation of 2-Arylimidazo[1,2-A]Pyridine with Cinnamalde-Hyde for Synthesis of N-Heterocycle
*Corresponding author: Nan Lu, College of Chemistry and Material Science, Shandong Agricultural University, China.
Received: April 08, 2024; Published: April 17, 2024
DOI: 10.34297/AJBSR.2024.22.002929
Abstract
The mechanism is investigated for regio-divergent oxidative annulation of 2-arylimidazo[1,2-a]pyridine with cinnamaldehyde. For Rh(III) catalyzed reaction, the coordination at N exists for initial complex followed by C-H activation of phenyl and chelation. Seven-membered rhodacycle generated by rollover undergoes elimination reduction. For Pd(II) catalyzed case, the palladation at imidazolyl C takes place followed by alkenylated insertion and metalation deprotonation furnishing six-membered palladacycle, which experiences reductive elimination. The olefin insertion is rate-limiting for both Rh(III) and Pd(II). The regio-divergence of dominant product is kinetically controlled by catalyst presenting as six-membered N-heterocycle preferred by Rh(III) and fivemembered isomer favored by Pd(II). The promotion of catalyst is in the deprotonation assisted by ligand and coordination of metal. The positive solvation effect was suggested by decreased relative and activation energies in DMA and DMF solution. These results are supported by Frontier MO analysis on specific TSs, ELF isosurface, and MBO value of vital bonding, breaking.
Keywords: Oxidative annulation, Regio-divergent, Isomeric N-heterocycle, Olefin insertion, Reductive elimination
Introduction
As an interesting part with privileged chemical moiety, imidazo[1,2-a]pyridines have been found in several marketed drugs. The necopidem and saripidem were used for treatment of anxiety and insomnia [1]. The therapeutic effect of minodronic acid was evaluated for osteoporosis [2]. Olprinone was a newly developed phosphodiesterase III inhibitor and GSK812397 was CXCR4 receptor antagonist [3,4]. Hence, the bicyclic system imidazo-[1,2-a]pyridine with bridgehead nitrogen was an important biologically active moiety towards synthetic drug molecule in medicinal chemistry [5]. In addition, with extended π-system, this framework has shown electronic and optical properties [6,7]. The imidazo[1,2-a]pyridine has been targeted for synthetic chemists in decades due to its significance in material chemistry. Especially the C3-functionalized imidazo[1,2-a]pyridine was more easily attacked by electrophile or radical with electron-rich character including C3-alkylation, C3-carbonylation, C3-arylation, C3-selenation, C3-N-Substitution, and C3-halogenation [8]. Recently, transition-metal-catalyzed regioselective functionalization and transition-metal-free C-H functionalization have attracted considerable attention [9,10]. Therefore, it is important to construct such molecular skeleton efficiently and greenly.
As versatile organic electronic material, arene and heteroarene have received considerable attention with extended conjugated π-system [11]. These hybrid scaffolds especially 2-arylimidazo[1,2-a]pyridine were key components of pharmaceuticals [12]. The annulation with appropriate coupling partner catalyzed by transition-metal could lead to several heteropolycyclic aromatic systems. The direct dehydrogenative annulation of imidazo[1,2-a]pyridines with diarylalkynes catalyzed by Pd(OAc)2 was developed to synthesize π-conjugated polyaromatic heterocycles [13]. Rh(III)-catalyzed annulative C-H functionalization of arenes with sulfoxonium ylides was realized affording structurally diverse fused hetero- and carbocycles [14]. The naphtho[1′,2′:4,5]imidazo[1,2-a]-pyridine was synthesized from cascade reaction of 2-arylimidazo[1,2-a]pyridines with α-diazo carbonyl compounds via Rh(III)-catalyzed regioselective C(sp2)-H alkylation followed by intramolecular annulation [15]. The bridged imidazopyridine was yielded by Rh(III)-catalyzed bicyclization with cyclic enone [16]. A selenium-coordinated Pd(II) trans-dichloride could catalyze site-selective annulation of 2-arylimidazo[1,2-a]pyridines leading to 2,3,4-triarylphenyl-1,7b-diaza-cyclopenta[cd]indene [17]. The naphtho[1′,2′:4,5]imidazo[1,2-a]pyridine was produced via oxidative C-H/C-H annulation of imidazopyridine with vinylene carbonate catalyzed by Rh(III) [18]. The Rh(III)-catalyzed [4 + 2] oxidative cycloaddition was reported to furnish maleimide fused benzocarbazoles [19]. Similarly, a diverse range of benzo[e]pyrido[1',2':1,2]imidazo[4,5-g]isoindole was achieved through Ru(II)-catalyzed C-H metalation followed by maleimide insertion and intramolecular cyclization [20].
As powerful platform of molecular synthesis, many advances are developed in electro catalytic, photo induced and asymmetric C-H activation [21]. Therefore the progress of transition-metal-catalyzed C-H activation in heterocycle has been particularly noticeable in recent years, such as the access to isogranulatimide alkaloid via Rh(III)-catalyzed dehydrogenative annulation/spirocyclization and switchable regioselective hydroalkylation of 2-arylindole with maleimide [22,23]. After Hanchate reported the synthesis of furanone-fused 1,2-benzothiazine via Rh(III)-catalyzed sulfoximine-directed C-H activation [24], a new breakthrough was Meena’s construction of fused N-heterocycle via regioselective oxidative annulation of 2-arylimidazo[1,2-a]pyridine with cinnamaldehyde [25]. Although two different isomeric annulated products were provided, there is no report about detailed mechanistic study explaining the obvious advantage of regioselective oxidative annulation. What’s the function of [RhCp*Cl2]2 in the generation of 5-arylnaphtho-[1′,2′:4,5]imidazo[1,2-a]pyridine-6-carbaldehyde?
How 1,7-diarylimidazo[5,1,2-cd]indolizine-6-carbaldehyde was afforded with Pd(OAc)2 as catalyst? Why the regioselectivity was dependent on different transition-metal, wherein the regulation of changes in valence states and the effects of solvent? To solve these mechanic problems in experiment, an in-depth theoretical study was necessary for this strategy applied versatile 2-arylimidazo[1,2-a]pyridine. The Density Functional Theory (DFT) method was employed focusing on the promotion during the transformation from Rh(III) to Rh(I) and from Pd(II) to Pd(0).
Computational Details
The geometry optimizations were performed at the B3LYP/BSI level with the Gaussian 09 package [26,27]. The mixed basis set of LanL2DZ for Rh, Pd and 6-31G(d) for non-metal atoms [28-32] was denoted as BSI. Different singlet and multiplet states were clarified with B3LYP and ROB3LYP approaches including Becke's three-parameter hybrid functional combined with Lee-Yang-Parr correction for correlation [33-39]. The nature of each structure was verified by performing harmonic vibrational frequency calculations. Intrinsic Reaction Coordinate (IRC) calculations were examined to confirm the right connections among key transition-states and corresponding reactants and products. Harmonic frequency calculations were carried out at the B3LYP/BSI level to gain Zero-Point Vibrational Energy (ZPVE) and thermodynamic corrections at 393 K, 413 K and 1 atm for each structure in N,N-dimethylacetamide (DMA) and N,N-dimethylformamide (DMF). The solvation-corrected free energies were obtained at the B3LYP/6-311++G(d,p) (LanL2DZ for Rh, Pd) level by using Integral Equation Formalism Polarizable Continuum Model (IEFPCM) in Truhlar’s “density” solvation model [40-42] on the B3LYP/BSI-optimized geometries.
As an efficient method obtaining bond and lone pair of a molecule from modern ab initio wave functions, NBO procedure was performed with Natural bond orbital (NBO3.1) to characterize electronic properties and bonding orbital interactions [43-44]. The representation of electron localization function (ELF) attractors were obtained through Natural Population Analysis (NPA) based on the Molecular Electron Density Theory (MEDT) [45]. The wave function analysis was provided using Multiwfn_3.7_dev package [46] including research on Frontier Molecular Orbital (FMO), ELF and Mayer Bond Order (MBO).
Results and Discussion
Based on previous research [21-25], the mechanism was explored for regioselective oxidative annulation of 2-arylimidazo[1,2-a]pyridine 1 with cinnamaldehyde 2 catalyzed by and leading to different isomeric N-heterocycle 3 (path A) and 4 (path B) (Scheme 1). Illustrated by red arrow of Scheme 2, the monomeric cationic rhodium complex Cp*RhCl2 was taken as model catalyst of Rh(III) to coordinate with N atom of 1 forming intermediate A. After the removal of HCl, a rhodacycle B was generated via o-C-H bond activation of C1-phenyl ring. The chelation of it with 2 afforded C followed by oxidative migratory insertion into Rh-C1 bond. The elimination of another HCl promoted rollover along C5-C6 axis producing seven-membered rhodacycle D which underwent reduction giving E and Cp*Rh species. Finally, oxidation of E and Rh(I) furnished six-membered annulated product 3 and recovered Rh(III).
In the case catalyzed by PdL2(OAc)2 as Pd(II) (blue arrow), the electrophilic palladation at imidazolyl C4 of 1 deliver-ing HOAc takes place initially to generate intermediate I, from which the insertion of 2 afforded the carbopalladation II. Then the alkenylated III was produced via β-hydrogen elimination with the help of OAc and Pd(0)L2. Subsequently, IV was obtained with palladation C7 which furnished a six-membered palladacycle V by concerted metalation deprotonation. The reductive elimination from V yielded five-membered annulated product 4 and the regeneration of Pd(II) upon oxida-tion of Pd(0). The optimized structures of TSs in Scheme 2 were listed by Figure 1. The activation energy was shown in Table 1 for all steps. Supplementary Table S1, Table S2 provided the relative energies of all stationary points. According to experiment, the Gibbs free energies in DMA and DMF solution phase are discussed here.
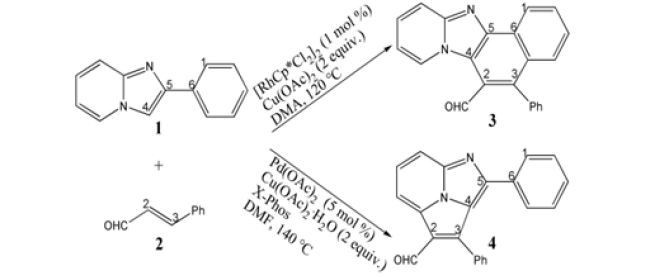
Scheme 1: Regioselective oxidative annulation of 2-arylimidazo[1,2-a] pyridine 1 with cinnamaldehyde 2 catalyzed by Rh(III) and Pd(II) leading to different isomeric N-heterocycle 3 and 4.

Scheme 2: Proposed reaction mechanism of regioselective oxidative annulation of 1 with 2 leading to 3 preferred by Rh(III) and 4 favored by Pd(II). TS is named according to the two intermediates it connects.
Rh(III)-catalyzed Annulation Via Path A
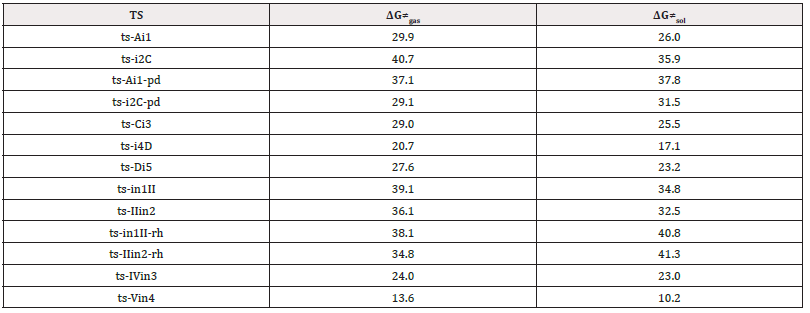
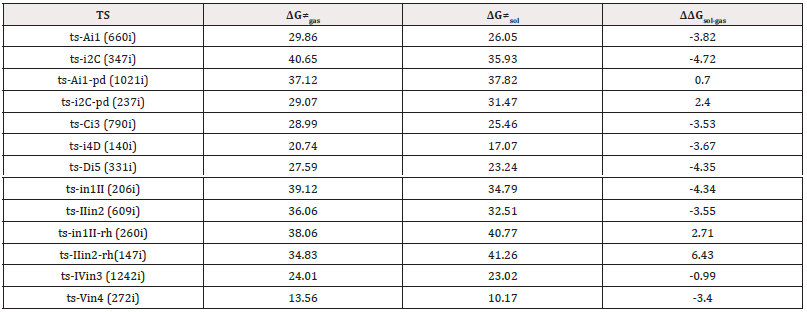
Initially, a complex A was located through the coordination of Cp*RhCl2 with 1 at N atom (black dash line of Figure 2a). With the reactive intermediate in hand, dehydrogenation proceeds via ts-Ai1 as step 1 with a mediate activation energy of 26.0 kcal mol-1 relative to the starting point A. The formation of i1 realized the o-C-H bond activation at C1-phenyl ring endoergic by 15.8 kcal mol-1. The transition vector of ts-Ai1 corresponds to the moving of H1 from C1 to Cl1 and closing of C1 to Rh slightly later (1.32, 1.67, 2.19 Å) (Figure S1a). This indicated the activation of phenyl C1 was driven by one coordinating Cl atom of catalyst and the Rh···C1 bonding was further promoted by Rh(III). After the removal of HCl, a five-membered rhodacycle B was generated involving a similar relative energy with i1. Thankfully the following combination of 2 and B brought about the 7.7 kcal mol-1 energy decrease of i2 as the starting point of step 2, which occurs via ts-i2C with activation energy of 35.9 kcal mol-1 endoergic by 1.6 kcal mol-1 furnishing seven-membered complex C with Rh-C2, C2-C3 and C1-C3 single bonds. The transition vector includes the elongation of Rh-C1, C2-C3 and concerted approach of Rh···C2, C1···C3 (2.24, 1.46, 2.15 and 1.98 Å), suggesting oxidative migratory insertion of C2-C3 double bond into Rh-C1 (Figure S1b). Kinetically, this chelation process is determined to be rate-limiting of path A leading to product 3.
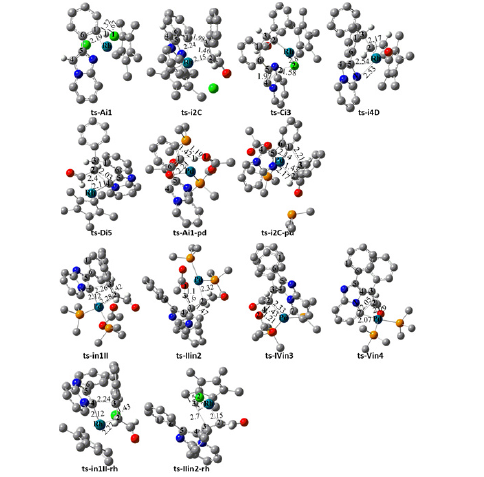
Figure 1: Proposed reaction mechanism of regioselective oxidative annulation of 1 with 2 leading to 3 preferred by Rh(III) and 4 favored by Pd(II). TS is named according to the two intermediates it connects.
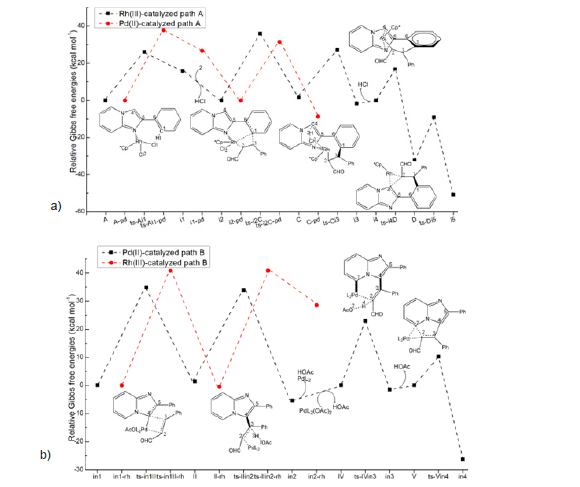
Figure 2: Relative Gibbs free energy profile catalysed by Rh(III) and Pd(II) in solvent phase starting from complex (a) A, A-pd, i2, i2-pd, i4 (b) in1, in1-rh, IV, V.
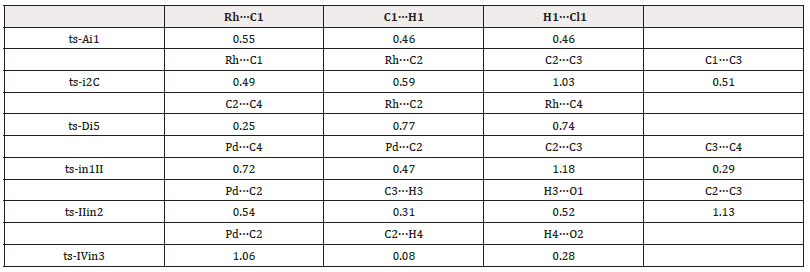
To highlight the idea of feasibility for changes in electron density and not molecular orbital interactions are responsible of the reactivity of organic molecules, quantum chemical tool Multiwfn was applied to analyze of electron density such as topological analysis of ELF, MBO results of bonding atoms and contribution of atomic orbital to HOMO of typical TSs (Table 2,3). ELF isosurface was presented by Figure 3 with green color denoting the electron localization. The marked small green part means good electron delocalization between two bonding atoms. The progress of bonding changes were fo-cused on along reaction coordinate based on a new reactivity theory named MEDT. On ELF isosurface of ts-Ai1, besides obvious connection of Rh-C1, the electron is mainly localized on H1 and Cl1 in agreement with the d electron of Rh (23.49%), bonding orbital of C1-H1 (6.77%, 3.34%) and p electron of Cl1 (7.07%) contributed to HOMO (Figure 4). This is beneficial for the abstraction of H1 by Cl1 and Rh-C1 bonding just as the results of MBO values for H1···Cl1 and Rh···C1 (0.46, 0.55). The correctness of ts-i2C is also verified by ELF analysis and HOMO composition. There is direct Rh-C2 linkage (25.56%, 3.23%) and bonding electron between C1 and C3 (5.61%, 8.86%), which is confirmed by MBO values of Rh···C2 and C1···C3 (0.59, 0.51).
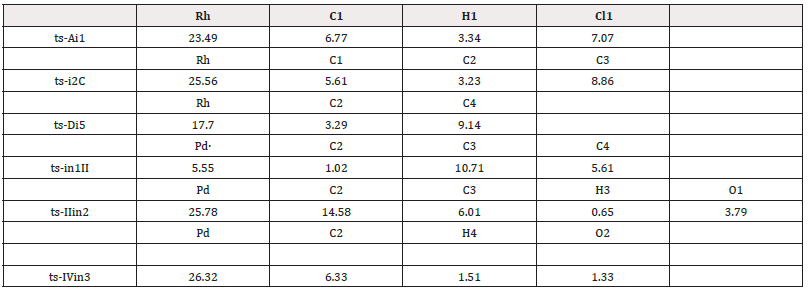
Table 3: Contribution (%) of Natural Atomic Orbital (NAO) to Highest Occupied Molecular Orbital (HOMO) of typical transition states.

Figure 3: Representation of ELF graph (isosurface value=0.84), attractors and valence basin population for typical TSs.

Figure 4: Highest Occupied Molecular Orbital (HOMO) of typical TSs. Different colors are used to identify the phase of the wave functions.
The next cleavage of another coordinated Cl atom in forms of HCl occurs in step 3 to get ready for the coordination of Rh transfer from N to C4 of step 4. Via ts-Ci3, the activation energy is 25.5 kcal mol-1 with respect to C exothermic by -1.7 kcal mol-1 leading to i3, from which the rupture of HCl gives i4 as the new starting point of following ring rotation. With previous preparation the rollover along C5-C6 axis is fairly easy via ts-i4D with a smaller barrier of 17.1 kcal mol-1 tremendously exothermic by -32.5 kcal mol-1 delivering stable complex D with seven-membered rhodacycle. Besides the breaking of Cl2 from Rh (2.8 Å), the transition vector of ts-Ci3 corresponds to the shift of H2 from C4 to Cl2 (1.97, 1.58 Å). The transition vector of ts-i4D indicates a loose structure with closing of Rh to C5 and transient no longer boned of Rh···C2 (2.54, 2.17 Å). The above two steps are both readily accessible with low activation energy from kinetics and quite favorable from thermodynamics with huge heat release.
From D the last step 5 takes place via ts-Di5 with small barrier of 23.2 kcal mol-1 continuously exothermic by -50.9 kcal mol-1. When this reduction elimination was completed, i5 would be generated binding complex E and Cp*Rh as precursors of one annulated product 3 and recovered catalyst Rh(III) with the participation of oxidants. From the transi-tion vector of ts-Di5, C2 and C4 is linking to form new six-membered ring along with the stretching of Rh-C2, Rh-C4 bond to squeeze Rh (2.03, 2.40, 2.11 Å) (Figure. S1c). The rationality of ts-Di5 can be approved by ELF isovalue of MEDT analy-sis. Although Rh-C2, Rh-C4 single bond still exists, there is remarkable green color denoting electron distribution between C2 and C4 (3.29%, 9.14%). The upcoming formation of C2-C4 bond is in accordance with MBO values for Rh···C2, Rh···C4, C2···C4 (0.77, 0.74, 0.25).
Pd(II)-Catalyzed Annulation Via Path B
When the reaction was catalyzed by Pd(II), the process delivering another major isomeric annulated product 4 was lo-cated as path B (black dash line of Figure 2b). The initial electrophilic palladation forms C4-Pd bond for complex I, which is reactive with a relative energy higher by 15.0 kcal mol-1 than isolated species of 1 and catalyst after the departure of HOAc. The introduction of 2 stabilizes intermediate in1 as the starting point, from which the insertion of olefin double bond takes place via ts-in1II in step 1 with a barrier of 34.8 kcal mol-1 generating carbopalladation complex II endoergic by 1.4 kcal mol-1. The transition vector corresponds to the stretching of C4-Pd to breaking, extension of C2-C3 double bond to single one and the simultaneous bonding of C2···Pd, C3···C4 (2.12, 1.42, 2.28 and 2.26 Å) (Figure S1d). Next step 2 proceeds via ts-IIin2 with a slightly lower barrier of 32.5 kcal mol-1 relative to II exothermic by -5.4 kcal mol-1 affording stable intermediate in2. Thus, step 1 is determined to be rate-limiting of path B, which is also the insertion process of 2. Two aspects of atomic motions can be seen from the transition vector. One is H3 transferring from C3 to O1 (1.60, 1.10 Å) denoting β-hydrogen elimination assisted by OAc. The other is C2-Pd fracture and C2-C3 contracted to double one (2.32, 1.47 Å). The alkenylated III is produced after the catalyst leaving in forms of HOAc and and Pd(0)L2.
The complex IV with C7-Pd bond is formed through a second electrophilic palladation based on the reintroduction of PdL2(OAc)2 and the removal of one HOAc just like the generation of I. The step 3 occurs via ts-IVin3 from IV with acti-vation energy of 23.0 kcal mol-1 furnishing intermediate in3 exothermic by -1.5 kcal mol-1. The transition vector demon-strates a concerted metalation deprotonation in collaborative asynchronous mode including the earlier H shifting C2···H4···O2 and later linkage of C2-Pd (1.43, 1.21, 2.33Å) (Figure S1f). Evidently, H4 can be easily taken away from C2 driven by OAc of catalyst. This assembles HOAc on one hand to leave freely and makes C2 more negative enhancing its nucleophilicity to Pd on the other. Hence in3 is characterized with stable six-membered palladacycle and C2-C3 double bond, from which complex V is given after the departure of HOAc. The reductive elimination takes place in step 4 via ts-Vin4 with lower barrier of 10.2 kcal mol-1 more dramatically exothermic by -26.3 kcal mol-1 delivering in4. Involving ring closure via C2···C7 approaching and departure of C2···Pd, C7···Pd (2.05, 2.09, 2.07 Å), the transition vector presents a process squeezing Pd(0)L2 and liberating five-membered product 4.
On ELF isosurface of ts-in1II, both the linkage between C2 and Pd and green color between C3 and C4 are obvious denoting bonding interaction. This echoes MBO values of Pd···C2 and C3···C4 (0.47, 0.29). Meanwhile Pd is leaving C4 together with the elongation of C2···C3 (0.72, 1.18). Here HOMO is composed by d electron of Pd, p lone pair of C4 (5.55%, 5.61%) and anti-bonding orbital on C2-C3 (1.02%, 10.71%), which indicates the coordination on Pd changing from C4 to C2. For ts-IIin2, C2 and Pd is still bonded with MBO value of 0.54. H3 is already leaving from C3 to O1 shown by the green color on ELF isosurface among them as well as MBO values for C3···H3 and H3···O1 (0.31, 0.52). The mismatch of d orbital on Pd (25.78%) and anti-bonding π electron on C2-C3 (14.58%, 6.01%) predicts the cleavage of C2-Pd. The electron on p lone pair orbital of O1 (3.79%) is conducive for H3 to approach. When it comes to ts-IVin3, HOMO is mainly located on bonding orbital of C2-Pd (26.32%, 6.33%) beneficial for the linkage. A small part on H4 and O2 points the following bonding between them (1.51%, 1.33%). The correctness is verified by green bond on C2-Pd and color by ELF analysis. MBO values for Pd···C2 and H4···O2 (1.06, 0.28) is also according to this outcome.
Regio-Divergence and Solvent Effect
To explore the relation of catalysts with regio-divergence puzzled in experiment, the annulation via Pd(II)-catalyzed path A and Rh(III)-catalyzed path B are also investigated (red dash line of Figure 2) in contrast with dominant paths (black dash line).
On one hand for the former, the H1 capture was realized with the assistance of OAc and phenyl C1 was coordinated to Pd(II) via ts-Ai1-pd (1.42, 1.19, 2.21 Å) (Figure 1) with a barrier of 37.8 kcal mol-1 relative to its starting point A-pd, which turns to be rate-limiting as step 1. The olefin insertion into Pd-C1 in step 2 is via ts-i2C-pd with a barrier of 31.5 kcal mol-1 with respect to i2-pd. Compared with the case of Rh(III), although the barrier is slightly reduced owing to one OAc ligand not bonded to Pd alleviating space congestion, path A is more preferred by Rh(III) considering the lower barrier of chela-tion in rate-limiting step 2 (35.9 kcal mol-1) yielding dominant 3 as regioselectivity. On the other for the latter, the barriers of step 1 via ts-in1II-rh and step 2 via ts-IIin2-rh (40.8, 41.3 kcal mol-1) are both higher distinctly than the case with Pd(II) involving rate-limiting step 1 (34.8 kcal mol-1). Furthermore from thermodynamics, compared with stable interme-diate in2 exothermic by -5.4 kcal mol-1, the production of in2-rh is required to be endoergic by 28.6 kcal mol-1. Hence the regioselectivity of 4 via path B dominated by Pd(II) is more apparently. Finally, the region-divergence of isomeric 3 and 4 are both kinetically controlled by catalyst presenting with six- and five-membered annulated cycles.
The impact of DMA and DMF solution is studied in view of the solvent effect on reaction estimated by our approach [28-32]. Obviously, the absolute energies of all stationary points in solution are lower than those in gas phase (Table S1). In general, DMA exerted bigger influence than DMF on annulation via both path A and B with relative energies de-creased by -21~-45 kcal mol-1 vs -20~-35 kcal mol-1. This range was -25~-45 kcal mol-1 vs -21~-27 kcal mol-1 in DMA for path A catalyzed by Rh(III) and Pd(II). For most steps, the activation energies are reduced in solution phase compared with in gas (Table S2). With reduction values -3~-5 kcal mol-1, the solvent effect of DMA is also more visible than DMF. Accordingly, the DMA and DMF solution both produce favorable influence on this regioselective oxidative annulation of 2-arylimidazo[1,2-a]pyridine with cinnamaldehyde catalyzed by Rh(III) and Pd(II) leading to different isomeric N-heterocycles.
Conclusions
Our DFT calculations provide the first theoretical investigation on regio-divergent oxidative annulation of 2-arylimidazo[1,2-a]pyridine with cinnamaldehyde. For reaction catalyzed by Rh(III), the coordination at N exists for initial complex followed by C-H activation of phenyl and chelation. The seven-membered rhodacycle generated by rollover undergoes elimination reduction gives six-membered N-heterocycle preferred as major product. For Pd(II) cata-lyzed case, the palladation at imidazolyl C takes place at first followed by alkenylated insertion and metalation deproto-nation furnishing six-membered palladacycle, the reductive elimination of which yields favored isomer with five-membered N-heterocycle. The olefin insertion is rate-limiting step for both Rh(III) and Pd(II).
The promotion of catalyst is reflected in the deprotonation assisted by ligand OAc or Cl and coordination of metal. Based on the comparison between possible paths, regio-divergence of isomeric product is kinetically controlled by cata-lyst presenting as six- and five-membered annulated cycles. The positive solvation effect is suggested by decreased abso-lute and activation energies in DMA and DMF solution compared with in gas. These results are supported by Multiwfn analysis on ELF isosurface, FMO composition of specific TSs, and MBO value of vital bonding, breaking.
Electronic Supplementary Material
Supplementary data available: [Computation information and cartesian coordinates of stationary points; Calculated rela-tive energies for the ZPE-corrected Gibbs free energies (ΔGgas), and Gibbs free energies (ΔGsol) for all species in solution phase at 393, 413 K.]
Acknowledgements
This work was supported by National Natural Science Foundation of China (21973056, 21972079) and Natural Science Foundation of Shandong Province (ZR2019MB050) and Key Laboratory of Agricultural Film Application of Ministry of Agriculture and Rural Affairs, P.R. China.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sanger DJ (1995) Behavioural effects of novel benzodiazepine (ω) receptor agonists and partial agonists: increases in punished responding and antagonism of the pentylenetetrazole cue. Behav Pharmacol 6(2): 116-126.
- Kitamura N, Shiraiwa, H, Inomata H, Nozaki T, Ikumi N, et al. (2018) Efficacy and safety of minodronic acid hydrate in patients with steroid-induced osteopo-rosis. Int J Rheum Dis 21: 813-820.
- Mizushige K, Ueda T, Yukiiri K, Suzuki H (2002) Olprinone: A phosphodiesterase III inhibitor with positive inotropic and vasodilator effects. Cardiovasc Drug Rev 20(3): 163-174.
- Jenkinson S, Thomson M, McCoy D, Edelstein M, Danehower S, et al. (2010) Blockade of X4-tropic HIV-1 Cellular entry by GSK812397, a potent noncompetitive CXCR4 receptor antagonist. Antimicrob. Agents Chemother 54: 817-824.
- Goel R, Luxami V, Paul K (2016) Imidazo[1,2-a] pyridines: Promising drug candidate for antitumor therapy. Curr Top Med Chem 16(30): 3590-3616.
- Kielesiński L, Tasior M, Gryko DT (2015) Polycyclic imidazo[1,2-a] pyridine analogs - synthesis via oxidative intra-molecular C-H amination and optical properties. Org Chem Front 2: 21-28.
- Firmansyah D, Banasiewicz M, Deperasińska I, Makarewicz A, Kozankiewicz B, et al. (2014) Vertically π-expanded imidazo[1,2-a] pyridine: The missing link of the puzzle. Chem Asian J 9(9): 2483-2493.
- Tashrifi Z, Mohammadi Khanaposhtani M, Larijani B, Mahdavi M (2020) C3-Functionalization of imid-azo[1,2-a] pyridines. Eur J Org Chem 2020: 269-284.
- Konwar D, Bora U (2021) Recent developments in transition-metal-catalyzed regioselective functionalization of imid-azo[1,2-a] pyridine. Chemistry Select 6: 2716-2744.
- Shi L, Li T, Mei GJ (2022) Recent advances in transition-metal-free C-H functionalization of imid-azo[1,2-a]-pyridines. Green Synth Catal 3: 227-242.
- Wang C, Dong H, Hu W, Liu Y, Zhu D (2012) Semiconducting π-Conjugated Systems in Field-Effect Transistors: A Material Odyssey of Organic Electronics. Chem Rev 112(4): 2208-2267.
- Samanta S, Mondal S, Ghosh D, Hajra A (2019) Rhodium-Catalyzed Directed C-H Amidation of Imidazoheterocy-cles with Dioxazolones. Org Lett 21(12): 4905-4909.
- Ghosh M, Naskar A, Mishra S, Hajra A (2015) Palladium-catalyzed dehydrogenative annulation of imid-azo[1,2-a] pyridines with diary-lalkynes. Tetrahedron Lett 56: 4101-4104.
- Zheng G, Tian M, Xu Y, Chen X, Li X (2018) Rhodium (III)-catalyzed annulative coupling between arenes and sul-foxonium ylides via C-H activation. Org Chem Front 5: 998-1002.
- Li B, Shen N, Zhang X, Fan X (2019) Synthesis of fused imidazo[1,2-a] pyridines derivatives through cascade C (sp 2) -H functionalizations. Org Biomol Chem 17: 9140-9150.
- Reddy KN, Chary DY, Sridhar B, Reddy BVS (2019) Rh (III)-catalyzed tandem bicyclization of 2-arylimidazo[1,2-a] pyridines with cyclic enones for the construction of bridged scaffolds. Org Lett 21: 8548-8552.
- Meena N, Sharma S, Bhatt R, Shinde VN, Sunda AP, et al. (2020) A seleni-um-coordinated palladium (II) trans-dichloride molecular rotor as a catalyst for site-selective annulation of 2-arylimidazo[1,2-a] pyridines. Chem Commun 56: 10223-10226.
- Ghosh K, Nishii Y, Miura M (2020) Oxidative C-H/C-H annulation of imidazopyridines and indazoles through rhodium-catalyzed vinylene transfer. Org Lett 22: 3547-3550.
- Li B, Guo C, Shen N, Zhang X, Fan X (2020) Synthesis of maleimide fused benzocarbazoles and imid-azo[1,2-a] pyridines via rhodium (III)-catalyzed [4 + 2] oxidative cycloaddition. Org Chem Front 7: 3698-3704.
- Yogananda Chary D, Nagarjuna Reddy K, Sridhar B, Subba Reddy BV (2021) Ru (II) catalyzed dehydrogenative annulation of 2-arylimidazo[1,2-a] pyridines with maleimides. Tetrahedron Lett 66: 152830.
- Rogge T, Kaplaneris N, Chatani N, Kim J, Chang S, et al. (2021) C-H activation. Nat Rev Methods Primers 1: 43.
- Shinde VN, Rangan K, Kumar D, Kumar A (2021) Rhodium (III)-catalyzed dehydrogenative annulation and spirocyclization of 2-arylindoles and 2-(1H-pyrazol-1-yl)-1H-indoles with maleimides: A facile access to isogranulatimide alkaloid analogues. J Org Chem 86(3): 2328-2338.
- Nipate DS, Shinde VN, Rangan K, Kumar A (2021) Switchable regioselective hydroalkylation of 2-arylindoles with maleimides. Org Biomol Chem 19: 4910-4921.
- Hanchate V, Kumar A, Prabhu KR (2019) Synthesis of Furanone-Fused 1,2-Benzothiazine by Rh (III)-Catalyzed C-H Activation: Regioselective Oxidative Annulation Leading to in Situ Lactonization in One Pot. J Org Chem 84(17): 11335-11342.
- Meena N, Shinde VN, Swami PN, Rangan K, Kumar A, et al. (2023) Catalyst-Controlled Regiodivergent Oxidative An-nulation of 2-Arylimidazo[1,2-a] pyridines with Cinnamaldehyde Derivatives for Construction of Fused N-Heterocyclic Frameworks. J Org Chem 88(18): 12902-12913.
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al. (2010) Gaussian 09 (Revision B.01), Gaussian, Inc. Wallingford CT.
- Hay PJ, Wadt WR (1985) Ab initio effective core potentials for molecular calculations-potentials for the transi-tion-metal atoms Sc to Hg J Chem Phys 82, 270-283.
- Lv H, Han F, Wang N, Lu N, Song Z, et al. (2022) Ionic Liquid Catalyzed C-C Bond Formation for the Synthesis of Polysubstituted Olefins. Eur J Org Chem e202201222.
- Zhuang H, Lu N, Ji N, Han F, Miao C (2021) Bu4NHSO4‐Catalyzed Direct N‐Allylation of Pyrazole and its Deriva-tives with Allylic Alcohols in Water: A Metal‐free, Recyclable and Sustainable System. Advan Synthe Catal 363: 5461-5472.
- Lu N, Lan X, Miao C, Qian P (2020) Theoretical investigation on transformation of Cr (II) to Cr (V) complexes bearing tetra-NHC and group transfer reactivity. Int J Quantum Chem 120: e26340.
- Lu N, Liang H, Qian P, Lan X, Miao C (2020) Theoretical investigation on the mechanism and enantioselectivity of organocatalytic asymmetric Povarov reactions of anilines and aldehydes. Int J Quantum Chem 120: e26574.
- Lu N, Wang Y (2023) Alloy and Media Effects on the Ethanol Partial Oxidation Catalyzed by Bimetallic Pt6M (M= Co, Ni, Cu, Zn, Ru, Rh, Pd, Sn, Re, Ir, and Pt). Comput Theoret Chem 1228: 114252.
- Becke AD (1996) Density-functional thermochemistry. IV. A new dynamical correlation functional and implications for exact-exchange mixing. J Chem Phys 104: 1040-1046.
- Lee CT, Yang WT, Parr RG (1998) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter 37(2): 785-789.
- Catellani M, Mealli C, Motti E, Paoli P, Perez Carren˜o, et al. (2002) Palladium-Arene Interactions in Cat-alytic Intermediates: An Experimental and Theoretical Investigation of the Soft Rearrangement between η1 and η2 Co-ordination Modes. J AM CHEM SOC 124(16): 4336-4346.
- Zicovich Wilson CM, Pascale F, Roetti C, Saunders VR, Dovesi R, et al. (2004) Calculation of the Vibration Frequencies of α-Quartz: The Effect of Hamiltonian and Basis Set. J Comput Chem 25(15): 1873-1881.
- Nielsen RJ, Goddard WA (2006) Mechanism of the Aerobic Oxidation of Alcohols by Palladium Complexes of N-Heterocyclic Carbenes. J AM CHEM SOC 128(30): 9651-9660.
- Zandler ME, D’Souza F (2006) The remarkable ability of B3LYP/3-21G(*) calculations to describe geometry, spectral and electrochemical properties of molecular and supramolecular porphyrin-fullerene conjugates. 9(7-8): 960-981.
- Marenich AV, Cramer CJ, Truhlar DG (2009) Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J Phys Chem B 113(18): 6378-6396.
- Tapia O (1992) Solvent effect theories: Quantum and classical formalisms and their applications in chemistry and biochemistry. J Math Chem 10: 139-181.
- Tomasi J, Persico M (1994) Molecular Interactions in Solution: An Overview of Methods Based on Continuous Distributions of the Solvent. Chem Rev 94: 2027-2094.
- Tomasi J, Mennucci B, Cammi R (2005) Quantum Mechanical Continuum Solvation Models. Chem Rev 105(8): 2999-3093.
- Reed AE, Weinstock RB, Weinhold F (1985) Natural population analysis. J Chem Phys 83: 735-746.
- Reed AE, Curtiss LA, Weinhold F (1988) Intermolecular interactions from a natural bond orbital donor-acceptor viewpoint. Chem Rev 88: 899-926.
- Domingo LR, Ríos Gutiérrez M, Pérez P (2016) A new model for CeC bond formation processes derived from the Molecular Electron Density Theory in the study of the mechanism of [3+2] cycloaddition reactions of carbenoid nitrile ylides with electron-deficient ethylenes. Tetrahedron 72(12): 1524-1532.
- Lu T, Chen F (2012) Multiwfn: A multifunctional wavefunction analyzer. J Comput Chem 33(5): 580-592.
Supplementary Information
Theoretical Investigation on Rh(III)- And Pd(II)-Catalyzed Regioselective Oxidative Annulation of 2-Arylimidazo[1,2-A]Pyridine with Cinnamaldehyde for Synthesis of N-Heterocycle
Nan Lu*, Chengxia Miao and Xiaozheng Lan
College of Chemistry and Material Science, Shandong Agricultural University, China.
Table of Contents Pages
Computation information and geometries of stationary points; 2-22
Additional tables and figures 23-26
Software: GAUSSIAN09
Level of Theory: B3LYP
Basis Set: 6-31G(d)
Geometry [Cartesian coordinates]:
Optimized Cartesian coordinates for ts-Ai1
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 1.385763 1.745614 -0.858810
2 6 0 0.456002 2.630505 -1.441679
3 6 0 0.868122 3.881518 -1.836412
4 6 0 2.220402 4.283223 -1.660849
5 6 0 3.124372 3.423456 -1.104109
6 6 0 3.423767 1.139439 -0.140578
7 6 0 2.516737 0.119658 0.057350
8 1 0 -0.563025 2.296009 -1.565629
9 1 0 0.159540 4.569351 -2.285215
10 1 0 2.550230 5.269301 -1.965819
11 1 0 4.169195 3.656576 -0.940972
12 1 0 4.472271 1.225910 0.092464
13 6 0 2.643592 -1.208640 0.625819
14 6 0 1.409004 -1.816572 0.998489
15 6 0 3.863113 -1.863268 0.805500
16 6 0 1.467077 -3.133141 1.506510
17 1 0 0.839324 -1.002406 1.873392
18 6 0 3.877736 -3.154027 1.330466
19 1 0 4.793287 -1.385974 0.509574
20 6 0 2.679915 -3.791761 1.677439
21 1 0 0.552301 -3.626788 1.818302
22 1 0 4.824194 -3.669980 1.464449
23 1 0 2.701852 -4.798030 2.086122
24 7 0 1.267991 0.503155 -0.382643
25 7 0 2.715352 2.170778 -0.715924
26 6 0 -2.175389 -0.384481 0.496443
27 6 0 -2.302415 -0.328809 -0.951646
28 6 0 -2.112776 -1.634867 -1.465108
29 6 0 -1.849720 -2.528607 -0.346874
30 6 0 -1.985011 -1.759345 0.864858
31 6 0 -2.420247 0.743254 1.451788
32 1 0 -3.457845 0.721021 1.811630
33 1 0 -1.747909 0.675029 2.310827
34 1 0 -2.255369 1.713907 0.977341
35 6 0 -2.003252 -2.283206 2.266280
36 1 0 -3.025310 -2.219133 2.661847
37 1 0 -1.703037 -3.332045 2.310928
38 1 0 -1.352137 -1.697817 2.923217
39 6 0 -1.709944 -4.015625 -0.475525
40 1 0 -2.686057 -4.482534 -0.661836
41 1 0 -1.044261 -4.267206 -1.304838
42 1 0 -1.296379 -4.461586 0.430988
43 6 0 -2.208773 -2.064369 -2.894218
44 1 0 -2.330837 -1.211474 -3.565000
45 1 0 -1.304696 -2.602749 -3.193298
46 1 0 -3.071422 -2.729322 -3.028023
47 6 0 -2.720259 0.865453 -1.757568
48 1 0 -2.156883 0.947102 -2.690770
49 1 0 -3.782138 0.785500 -2.023351
50 1 0 -2.603880 1.793918 -1.194560
51 45 0 -0.186440 -1.108610 -0.329745
52 17 0 1.118469 -2.113336 -2.151921
53 17 0 0.589268 -0.042525 3.225081
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-i2C
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 3.616071 -2.693174 2.804684
2 6 0 4.207175 -3.145880 1.606172
3 6 0 5.556435 -3.415635 1.603489
4 6 0 6.344340 -3.238941 2.773804
5 6 0 5.773611 -2.764467 3.920486
6 6 0 3.634481 -1.969694 4.933733
7 6 0 2.366119 -1.882284 4.402158
8 1 0 3.599919 -3.166593 0.694433
9 1 0 6.028982 -3.742635 0.683141
10 1 0 7.406861 -3.454593 2.766801
11 1 0 6.308322 -2.573678 4.842592
12 1 0 4.038236 -1.689387 5.893001
13 6 0 1.114757 -1.393111 4.959304
14 6 0 0.073158 -1.111928 4.038257
15 6 0 0.900533 -1.257336 6.337658
16 6 0 -1.174099 -0.718146 4.556248
17 6 0 -0.343196 -0.856975 6.821571
18 1 0 1.700995 -1.495512 7.033176
19 6 0 -1.388820 -0.597739 5.928169
20 1 0 -1.981420 -0.473942 3.869489
21 1 0 -0.500098 -0.752010 7.891010
22 1 0 -2.361136 -0.288110 6.301024
23 7 0 2.352904 -2.341002 3.104281
24 7 0 4.425983 -2.488130 3.930182
25 6 0 -0.700521 -4.100211 0.945721
26 6 0 0.404108 -4.710909 1.604512
27 6 0 0.082808 -4.842011 3.035387
28 6 0 -1.185578 -4.268162 3.243694
29 6 0 -1.650623 -3.723246 1.971064
30 6 0 -0.830858 -3.944524 -0.535961
31 1 0 -1.130224 -4.907165 -0.973110
32 1 0 -1.575402 -3.197224 -0.807581
33 1 0 0.124251 -3.636639 -0.978510
34 6 0 -3.032022 -3.189970 1.725575
35 1 0 -3.732186 -4.022900 1.576288
36 1 0 -3.398301 -2.605511 2.574040
37 1 0 -3.068864 -2.564337 0.832354
38 6 0 -1.962698 -4.234041 4.523898
39 1 0 -2.727992 -5.020966 4.521359
40 1 0 -1.321685 -4.396512 5.393045
41 1 0 -2.474525 -3.278693 4.660850
42 6 0 0.939830 -5.552862 4.038710
43 1 0 0.824957 -6.640409 3.943935
44 1 0 1.998391 -5.324568 3.889130
45 1 0 0.678111 -5.279046 5.063607
46 6 0 1.582921 -5.311330 0.908086
47 1 0 2.421221 -5.471360 1.589722
48 1 0 1.301466 -6.289347 0.493454
49 1 0 1.914612 -4.667555 0.084188
50 45 0 0.249774 -2.628493 2.401064
51 17 0 2.473213 -2.384789 -1.078819
52 6 0 0.228968 -0.849589 1.179663
53 6 0 0.451185 -0.069404 2.391797
54 1 0 1.020176 -1.031931 0.447939
55 1 0 -0.395947 0.566609 2.637760
56 6 0 1.738831 0.654998 2.610046
57 6 0 2.853048 0.465024 1.774841
58 6 0 1.801763 1.637570 3.613808
59 6 0 4.000249 1.239600 1.954968
60 1 0 2.828210 -0.268771 0.973139
61 6 0 2.947893 2.407402 3.790152
62 1 0 0.941228 1.794915 4.259681
63 6 0 4.054566 2.206897 2.959866
64 1 0 4.849420 1.088882 1.294978
65 1 0 2.976280 3.166829 4.566431
66 1 0 4.949151 2.809690 3.089943
67 6 0 -1.130828 -0.684713 0.629943
68 8 0 -1.501185 -0.921522 -0.506147
69 1 0 -1.869220 -0.287727 1.373894
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-Ai1-pd
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 -1.400705 3.408822 -3.412884
2 6 0 -0.249201 3.715590 -4.174320
3 6 0 0.019430 5.022394 -4.505636
4 6 0 -0.855205 6.064920 -4.086737
5 6 0 -1.970564 5.777015 -3.350113
6 6 0 -3.268930 3.906778 -2.302664
7 6 0 -3.023041 2.537572 -2.285386
8 1 0 0.392666 2.896384 -4.477706
9 1 0 0.900244 5.267273 -5.089984
10 1 0 -0.648905 7.096956 -4.345943
11 1 0 -2.676799 6.520257 -3.000076
12 1 0 -4.066866 4.508418 -1.898372
13 6 0 -3.843625 1.490002 -1.655328
14 6 0 -3.570000 0.117156 -1.891160
15 6 0 -4.927859 1.855456 -0.840072
16 6 0 -4.423204 -0.835302 -1.300383
17 1 0 -3.327054 -0.163770 -3.264975
18 6 0 -5.740835 0.887862 -0.255332
19 1 0 -5.135641 2.904306 -0.645610
20 6 0 -5.496462 -0.468080 -0.489401
21 1 0 -4.241440 -1.891458 -1.486559
22 1 0 -6.570085 1.192852 0.377553
23 1 0 -6.136534 -1.225183 -0.044141
24 7 0 -1.872561 2.248375 -2.973974
25 7 0 -2.239070 4.471517 -3.017721
26 46 0 -1.441609 -0.512417 -1.898022
27 6 0 -2.162550 -0.831710 -4.753500
28 8 0 -3.312353 -0.381161 -4.439826
29 8 0 -1.220202 -1.024751 -3.935299
30 6 0 -1.924142 -1.139274 -6.213508
31 1 0 -1.861537 -0.195924 -6.765423
32 1 0 -0.997970 -1.698626 -6.345068
33 1 0 -2.770261 -1.700616 -6.616428
34 15 0 0.749898 -1.300196 -1.672032
35 15 0 -0.592984 -5.811807 1.275781
36 6 0 1.822489 -0.672543 -3.034827
37 1 0 2.811119 -1.142784 -3.014802
38 1 0 1.330226 -0.880918 -3.986526
39 1 0 1.936170 0.410242 -2.933753
40 6 0 1.695631 -0.880304 -0.153218
41 1 0 1.230211 -1.388094 0.692035
42 1 0 2.737772 -1.200854 -0.252560
43 1 0 1.660323 0.198726 0.014940
44 6 0 0.866067 -3.126684 -1.810561
45 1 0 0.405375 -3.440399 -2.750756
46 1 0 1.908448 -3.461516 -1.787630
47 1 0 0.316107 -3.573678 -0.979892
48 6 0 -2.385329 -5.344203 1.079010
49 1 0 -2.965447 -5.512802 1.994354
50 1 0 -2.426434 -4.282065 0.821211
51 1 0 -2.835755 -5.921433 0.265369
52 6 0 -0.777524 -7.490349 2.074400
53 1 0 -1.453380 -7.470894 2.937445
54 1 0 -1.164730 -8.206206 1.342363
55 1 0 0.202136 -7.850483 2.404432
56 6 0 -0.187091 -4.779730 2.772731
57 1 0 -0.861052 -4.979658 3.614436
58 1 0 0.842240 -4.968832 3.093527
59 1 0 -0.281427 -3.729738 2.480745
60 6 0 -1.482187 -0.937365 0.980138
61 8 0 -1.567929 -0.007160 0.079733
62 8 0 -1.125987 -2.108809 0.788438
63 6 0 -1.884532 -0.450309 2.369232
64 1 0 -1.472747 0.542340 2.567078
65 1 0 -2.975593 -0.365449 2.407410
66 1 0 -1.554615 -1.158460 3.130816
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-i2C-pd
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 0.393439 1.868566 -0.886176
2 6 0 1.492790 1.795415 -1.772821
3 6 0 2.197304 2.937653 -2.071093
4 6 0 1.824145 4.185416 -1.494851
5 6 0 0.754849 4.263318 -0.646720
6 6 0 -1.052925 2.918735 0.446151
7 6 0 -1.323384 1.558134 0.379840
8 1 0 1.740641 0.829412 -2.199937
9 1 0 3.043738 2.894805 -2.748677
10 1 0 2.381206 5.087350 -1.721137
11 1 0 0.417284 5.178532 -0.175618
12 1 0 -1.492326 3.720734 1.016670
13 6 0 -2.369543 0.827323 1.108787
14 6 0 -2.337101 -0.579655 1.268570
15 6 0 -3.414936 1.541753 1.720405
16 6 0 -3.320173 -1.210420 2.041386
17 6 0 -4.388321 0.901883 2.483964
18 1 0 -3.467927 2.619313 1.592261
19 6 0 -4.337010 -0.482344 2.659685
20 1 0 -3.285722 -2.289334 2.174048
21 1 0 -5.185344 1.483393 2.938609
22 1 0 -5.083453 -0.990412 3.264152
23 7 0 -0.430921 0.927085 -0.444054
24 7 0 0.044658 3.125067 -0.353859
25 6 0 -0.797376 -2.248214 -0.731704
26 6 0 -2.154502 -1.849835 -0.533273
27 1 0 -0.173186 -1.700802 -1.430573
28 1 0 -2.768114 -2.601458 -0.041968
29 6 0 -2.930617 -1.009069 -1.472497
30 6 0 -2.339113 -0.337318 -2.556016
31 6 0 -4.330291 -0.963639 -1.334867
32 6 0 -3.129578 0.361714 -3.467144
33 1 0 -1.264526 -0.359293 -2.684729
34 6 0 -5.116976 -0.267360 -2.246853
35 1 0 -4.796522 -1.468485 -0.493480
36 6 0 -4.517155 0.401214 -3.317512
37 1 0 -2.657633 0.875886 -4.299517
38 1 0 -6.195838 -0.245397 -2.123710
39 1 0 -5.128117 0.945618 -4.032003
40 6 0 -0.495681 -3.674136 -0.595018
41 8 0 0.463128 -4.243065 -1.111625
42 1 0 -1.221227 -4.252676 0.019079
43 46 0 -0.388600 -1.481089 1.263943
44 15 0 1.316224 -2.114056 -4.289582
45 15 0 1.822540 -2.326925 1.474290
46 6 0 -0.169061 -3.165121 -4.691013
47 1 0 0.025868 -3.866615 -5.510363
48 1 0 -0.441435 -3.734826 -3.798144
49 1 0 -1.017557 -2.529353 -4.961248
50 6 0 1.806795 -1.574446 -6.007368
51 1 0 2.768005 -1.052535 -5.967122
52 1 0 1.893736 -2.417013 -6.703255
53 1 0 1.064106 -0.872123 -6.398556
54 6 0 2.565954 -3.463893 -3.998855
55 1 0 2.278415 -4.008201 -3.094854
56 1 0 2.622070 -4.168667 -4.836581
57 1 0 3.557048 -3.027826 -3.839583
58 6 0 2.795619 -1.580484 2.843476
59 1 0 3.817892 -1.972244 2.841759
60 1 0 2.794747 -0.495344 2.729523
61 1 0 2.311064 -1.822147 3.792096
62 6 0 2.001516 -4.136060 1.796618
63 1 0 3.055407 -4.400300 1.933084
64 1 0 1.449472 -4.392461 2.704980
65 1 0 1.596220 -4.703076 0.956142
66 6 0 2.864973 -2.042753 -0.016124
67 1 0 3.882175 -2.420751 0.130114
68 1 0 2.404853 -2.552763 -0.865326
69 1 0 2.902007 -0.969846 -0.220816
70 6 0 0.260434 0.346033 3.474566
71 8 0 -0.297026 -0.809334 3.271523
72 8 0 0.891725 1.007072 2.642601
73 6 0 0.060368 0.870052 4.898638
74 1 0 0.753202 1.689311 5.099196
75 1 0 -0.965982 1.238900 5.001614
76 1 0 0.193361 0.071286 5.633202
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-Ci3
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 -5.289867 -3.036204 3.706420
2 6 0 -6.544617 -2.516711 4.108240
3 6 0 -6.837946 -2.466363 5.449253
4 6 0 -5.908832 -2.935287 6.424592
5 6 0 -4.698235 -3.418361 6.023925
6 6 0 -3.167139 -3.865841 4.147924
7 6 0 -3.418688 -3.719315 2.753449
8 1 0 -7.221174 -2.133691 3.358748
9 1 0 -7.785227 -2.048217 5.775428
10 1 0 -6.148783 -2.897472 7.480502
11 1 0 -3.915089 -3.776490 6.679717
12 1 0 -3.813514 -5.110206 2.758196
13 6 0 -2.284832 -3.621884 1.769478
14 6 0 -2.322215 -3.310577 0.388018
15 6 0 -1.017370 -3.846251 2.364637
16 6 0 -1.083970 -3.183205 -0.284695
17 6 0 0.173334 -3.746586 1.667905
18 1 0 -1.010747 -4.084764 3.421291
19 6 0 0.144188 -3.391254 0.318361
20 1 0 -1.106022 -2.929779 -1.341337
21 1 0 1.115117 -3.925362 2.178732
22 1 0 1.060281 -3.280577 -0.254599
23 7 0 -4.730640 -3.183433 2.502999
24 7 0 -4.386602 -3.461978 4.685345
25 6 0 -7.843377 -4.838486 0.427063
26 6 0 -7.906723 -3.390929 0.332825
27 6 0 -7.818454 -3.061693 -1.142503
28 6 0 -7.483953 -4.195380 -1.808054
29 6 0 -7.365889 -5.298029 -0.809602
30 6 0 -8.303523 -5.671836 1.596710
31 1 0 -7.832673 -6.656884 1.607532
32 1 0 -8.089813 -5.194584 2.558367
33 1 0 -9.391530 -5.815725 1.544015
34 6 0 -7.062370 -6.712820 -1.185852
35 1 0 -7.897356 -7.132830 -1.764694
36 1 0 -6.170916 -6.772665 -1.816057
37 1 0 -6.903296 -7.345519 -0.310746
38 6 0 -7.270174 -4.404000 -3.275419
39 1 0 -7.972509 -5.144069 -3.680890
40 1 0 -7.412295 -3.476278 -3.835029
41 1 0 -6.260097 -4.770961 -3.488458
42 6 0 -8.091996 -1.693655 -1.681527
43 1 0 -7.475019 -0.949623 -1.167132
44 1 0 -7.897387 -1.634299 -2.755447
45 1 0 -9.140620 -1.408097 -1.521235
46 6 0 -8.693270 -2.494667 1.245367
47 1 0 -8.206936 -1.518811 1.339244
48 1 0 -9.696521 -2.319809 0.831164
49 1 0 -8.824481 -2.932413 2.237342
50 45 0 -5.871136 -4.113080 0.996729
51 17 0 -4.665054 -6.326593 2.217944
52 6 0 -4.922563 -2.712312 -0.187085
53 6 0 -3.497440 -3.128048 -0.603481
54 1 0 -5.537470 -2.652755 -1.086434
55 1 0 -3.184704 -2.248525 -1.190577
56 6 0 -3.501898 -4.272194 -1.626164
57 6 0 -3.551138 -3.971125 -2.994525
58 6 0 -3.408355 -5.617350 -1.244056
59 6 0 -3.514867 -4.981058 -3.959095
60 1 0 -3.605653 -2.931511 -3.310967
61 6 0 -3.376016 -6.629631 -2.205475
62 1 0 -3.343654 -5.878554 -0.193219
63 6 0 -3.428121 -6.317688 -3.566462
64 1 0 -3.541628 -4.721004 -5.013812
65 1 0 -3.294948 -7.665149 -1.886450
66 1 0 -3.388037 -7.106621 -4.312150
67 6 0 -4.948532 -1.388304 0.525824
68 8 0 -5.888900 -0.609772 0.531916
69 1 0 -4.006031 -1.114875 1.043444
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-i4D
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 -0.683842 3.380178 1.466044
2 6 0 -0.355634 4.681452 1.917672
3 6 0 -0.501011 5.747319 1.061784
4 6 0 -0.982791 5.539526 -0.265190
5 6 0 -1.309608 4.283991 -0.693415
6 6 0 -1.459123 1.873967 -0.138138
7 6 0 -1.122044 1.297222 1.096763
8 1 0 -0.014795 4.812838 2.939754
9 1 0 -0.259559 6.752179 1.393754
10 1 0 -1.099072 6.376608 -0.944605
11 1 0 -1.685384 4.045657 -1.680520
12 6 0 -1.266743 -0.121035 1.508013
13 6 0 -0.692227 -1.234017 0.816854
14 6 0 -2.079705 -0.384806 2.649188
15 6 0 -1.027401 -2.538302 1.255833
16 6 0 -2.380920 -1.669340 3.053951
17 1 0 -2.493983 0.471178 3.171586
18 6 0 -1.860893 -2.763696 2.336148
19 1 0 -0.618962 -3.382471 0.706087
20 1 0 -3.033044 -1.834760 3.906974
21 1 0 -2.110636 -3.779633 2.629294
22 7 0 -0.659456 2.203882 2.086961
23 7 0 -1.168529 3.204731 0.147156
24 6 0 1.633045 1.069438 4.163869
25 6 0 1.527332 -0.327029 4.173577
26 6 0 2.445263 -0.866978 3.158775
27 6 0 3.212251 0.226120 2.627232
28 6 0 2.599276 1.433105 3.119849
29 6 0 0.870256 2.051561 4.996368
30 1 0 0.434877 1.573933 5.877296
31 1 0 1.524519 2.859477 5.340496
32 1 0 0.060080 2.485869 4.400771
33 6 0 3.052367 2.827316 2.820367
34 1 0 2.203944 3.513256 2.761116
35 1 0 3.724646 3.187943 3.610848
36 1 0 3.590357 2.872206 1.871257
37 6 0 4.499500 0.149616 1.869131
38 1 0 4.694332 -0.861914 1.506586
39 1 0 4.524494 0.815852 1.003595
40 1 0 5.320368 0.427761 2.542636
41 6 0 2.720970 -2.327841 2.968842
42 1 0 3.381767 -2.700393 3.763125
43 1 0 1.797593 -2.911756 3.006446
44 1 0 3.207558 -2.523797 2.010934
45 6 0 0.653460 -1.175555 5.042076
46 1 0 0.039209 -1.857947 4.445930
47 1 0 1.265016 -1.787289 5.717166
48 1 0 -0.021688 -0.569819 5.648807
49 45 0 1.082212 0.365666 1.970016
50 6 0 1.210532 0.099259 -0.180000
51 6 0 0.319324 -1.140228 -0.338660
52 1 0 0.805694 1.015800 -0.602370
53 1 0 0.987847 -1.994047 -0.161315
54 6 0 -0.263750 -1.385322 -1.733577
55 6 0 -1.230167 -0.543614 -2.305541
56 6 0 0.183457 -2.496807 -2.463897
57 6 0 -1.730800 -0.823455 -3.578223
58 1 0 -1.563180 0.338864 -1.753576
59 6 0 -0.320045 -2.772841 -3.736104
60 1 0 0.936886 -3.154650 -2.034220
61 6 0 -1.283952 -1.934536 -4.297152
62 1 0 -2.478219 -0.163533 -4.011438
63 1 0 0.040227 -3.639154 -4.284599
64 1 0 -1.680782 -2.144208 -5.286984
65 6 0 2.615189 -0.090479 -0.556341
66 8 0 3.381615 0.791470 -0.921365
67 1 0 2.989188 -1.140642 -0.458185
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-Di5
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 4.601376 -0.436689 5.026859
2 6 0 5.519803 -0.527377 6.095174
3 6 0 5.122021 -1.082037 7.289782
4 6 0 3.787821 -1.551852 7.451381
5 6 0 2.890093 -1.452383 6.424041
6 6 0 2.566564 -0.670472 4.049449
7 6 0 3.543021 -0.138565 3.188571
8 1 0 6.524835 -0.154847 5.932812
9 1 0 5.822681 -1.163883 8.114320
10 1 0 3.466258 -1.991119 8.388898
11 1 0 1.858358 -1.775238 6.482897
12 6 0 3.371230 0.116877 1.753999
13 6 0 2.561644 -0.713066 0.951896
14 6 0 4.065152 1.188169 1.160585
15 6 0 2.426995 -0.403966 -0.409782
16 6 0 3.927966 1.471874 -0.193502
17 1 0 4.713236 1.787703 1.791690
18 6 0 3.092100 0.677418 -0.983328
19 1 0 1.807665 -1.044619 -1.032993
20 1 0 4.465986 2.307718 -0.631633
21 1 0 2.969132 0.890293 -2.041409
22 7 0 4.754604 0.040820 3.794791
23 7 0 3.284581 -0.907582 5.225376
24 6 0 -1.599770 0.341065 4.571021
25 6 0 -0.712436 1.142748 5.436079
26 6 0 0.173056 1.839865 4.608018
27 6 0 -0.132540 1.468758 3.218206
28 6 0 -1.317335 0.645876 3.215235
29 6 0 -2.727681 -0.527316 5.047440
30 1 0 -3.663952 0.043606 5.107701
31 1 0 -2.887497 -1.366461 4.365059
32 1 0 -2.530507 -0.935350 6.042694
33 6 0 -2.045101 0.095070 2.024387
34 1 0 -3.098492 0.399920 2.028472
35 1 0 -1.603036 0.450912 1.090565
36 1 0 -2.011815 -1.002421 2.017434
37 6 0 0.482514 2.121961 2.018169
38 1 0 0.426779 1.487140 1.131548
39 1 0 -0.044965 3.059631 1.793568
40 1 0 1.533744 2.364457 2.180865
41 6 0 1.250452 2.803471 5.005796
42 1 0 0.982876 3.830767 4.726240
43 1 0 1.426049 2.789994 6.084118
44 1 0 2.197767 2.566848 4.511458
45 6 0 -0.819112 1.212596 6.930451
46 1 0 0.087193 1.622689 7.382244
47 1 0 -1.659301 1.852736 7.232259
48 1 0 -0.995328 0.227017 7.371472
49 45 0 0.467728 -0.467471 4.152223
50 6 0 1.690274 -2.151271 2.957767
51 6 0 1.853349 -1.975050 1.446928
52 1 0 2.447769 -2.771824 3.432210
53 1 0 0.829463 -1.925307 1.058431
54 6 0 2.468930 -3.265505 0.891919
55 6 0 1.640545 -4.381598 0.698917
56 6 0 3.839978 -3.385643 0.629520
57 6 0 2.171281 -5.587132 0.239227
58 1 0 0.581583 -4.297171 0.928447
59 6 0 4.369649 -4.593554 0.171944
60 1 0 4.492813 -2.529733 0.772156
61 6 0 3.537697 -5.696719 -0.027825
62 1 0 1.516225 -6.440998 0.089063
63 1 0 5.434187 -4.670091 -0.031930
64 1 0 3.951104 -6.634561 -0.388229
65 6 0 0.320117 -2.418745 3.419273
66 8 0 -0.642124 -2.914522 2.816018
67 1 0 0.285737 -2.360556 4.613407
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-in1II
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 2.924288 -0.227545 1.706101
2 6 0 3.852733 -1.012310 2.422520
3 6 0 3.620323 -2.360656 2.575361
4 6 0 2.452141 -2.950263 2.022187
5 6 0 1.546048 -2.180126 1.341097
6 6 0 1.011265 0.140631 0.532998
7 6 0 1.791035 1.318560 0.717771
8 1 0 4.727399 -0.521140 2.832920
9 1 0 4.324657 -2.977232 3.124361
10 1 0 2.259097 -4.010660 2.138803
11 1 0 0.617950 -2.543978 0.917161
12 6 0 1.572747 2.672417 0.179020
13 6 0 2.578445 3.644400 0.360915
14 6 0 0.279499 4.311117 -1.064658
15 6 0 2.431064 4.924887 -0.162868
16 1 0 3.468260 3.371155 0.916608
17 6 0 1.281222 5.264826 -0.882942
18 1 0 -0.623508 4.557951 -1.615605
19 1 0 3.216269 5.660761 -0.011540
20 1 0 1.168843 6.264478 -1.293717
21 7 0 2.929657 1.075290 1.429162
22 7 0 1.776831 -0.837822 1.185566
23 6 0 1.212554 -0.268368 -1.685767
24 6 0 0.018419 -0.979964 -1.979014
25 1 0 1.173383 0.801373 -1.879398
26 1 0 0.070244 -2.062922 -2.048265
27 46 0 -0.952951 -0.490732 0.028054
28 6 0 -3.312269 1.865964 -0.650235
29 8 0 -2.211817 1.521355 -1.182227
30 8 0 -3.838088 1.346937 0.370218
31 6 0 -4.043459 3.041567 -1.317255
32 1 0 -4.045084 2.921382 -2.404573
33 1 0 -5.066902 3.132560 -0.947357
34 1 0 -3.507581 3.972247 -1.095779
35 15 0 -2.875441 -1.914369 -0.530173
36 15 0 -1.565075 0.211381 2.224346
37 6 0 -3.823275 -2.733903 0.833209
38 1 0 -4.507538 -3.476877 0.411333
39 1 0 -3.144336 -3.235254 1.528697
40 1 0 -4.407153 -1.993087 1.379406
41 6 0 -4.227337 -1.199501 -1.548625
42 1 0 -3.804332 -0.806754 -2.474656
43 1 0 -4.968640 -1.970856 -1.782964
44 1 0 -4.680401 -0.384886 -0.980831
45 6 0 -2.366437 -3.395538 -1.520680
46 1 0 -1.578542 -3.951718 -1.003589
47 1 0 -3.222483 -4.059981 -1.678213
48 1 0 -2.000756 -3.050930 -2.490265
49 6 0 -0.411325 -0.340934 3.565268
50 1 0 -0.779907 0.007551 4.535537
51 1 0 -0.335650 -1.430955 3.585386
52 1 0 0.584842 0.073485 3.401234
53 6 0 -1.515885 2.041532 2.398163
54 1 0 -1.585338 2.307741 3.458353
55 1 0 -0.593561 2.445608 1.980308
56 1 0 -2.373787 2.439111 1.854876
57 6 0 -3.197221 -0.188388 2.980743
58 1 0 -3.292327 0.362886 3.922164
59 1 0 -3.970697 0.137988 2.284825
60 1 0 -3.279234 -1.255986 3.191710
61 6 0 2.562856 -0.840590 -1.698860
62 6 0 3.668515 0.034133 -1.702694
63 6 0 2.804410 -2.227463 -1.756803
64 6 0 4.967615 -0.459867 -1.772585
65 1 0 3.498308 1.104990 -1.652764
66 6 0 4.105508 -2.718220 -1.820122
67 1 0 1.973063 -2.923744 -1.770736
68 6 0 5.190517 -1.837755 -1.828720
69 1 0 5.805975 0.229967 -1.781853
70 1 0 4.274362 -3.789861 -1.870025
71 1 0 6.204138 -2.224000 -1.884074
72 6 0 -0.932507 -0.360969 -2.925943
73 8 0 -1.693754 -1.020087 -3.625275
74 1 0 -0.910956 0.739058 -2.983800
75 6 0 0.415174 3.025146 -0.537980
76 1 0 -0.403130 2.323508 -0.677182
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-IIin2
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 -2.790051 1.212953 2.827663
2 6 0 -3.686339 2.142166 3.407129
3 6 0 -4.598047 2.791504 2.610265
4 6 0 -4.642727 2.526959 1.211298
5 6 0 -3.780153 1.627574 0.647787
6 6 0 -1.882981 0.040721 1.118413
7 6 0 -1.287431 -0.252923 2.351812
8 1 0 -3.620700 2.311626 4.475868
9 1 0 -5.291376 3.505755 3.042693
10 1 0 -5.364717 3.028373 0.576859
11 1 0 -3.772416 1.367260 -0.402178
12 6 0 -0.218962 -1.217680 2.681506
13 6 0 0.693891 -0.901773 3.703491
14 6 0 0.847058 -3.396008 2.470636
15 6 0 1.675183 -1.812268 4.091543
16 1 0 0.602451 0.058668 4.199475
17 6 0 1.757907 -3.064608 3.475315
18 1 0 0.887683 -4.375512 2.001659
19 1 0 2.368341 -1.550515 4.886710
20 1 0 2.514569 -3.779858 3.785622
21 7 0 -1.835705 0.475312 3.378423
22 7 0 -2.866294 0.980359 1.444694
23 6 0 -1.607779 -0.268038 -0.319207
24 6 0 -0.937988 0.862149 -0.978193
25 1 0 -0.397791 -1.284923 -0.562728
26 1 0 -1.094791 1.830883 -0.513762
27 46 0 1.355862 0.612307 -0.701932
28 6 0 0.891635 -2.639490 -1.604734
29 8 0 0.623440 -1.546478 -0.858077
30 8 0 2.007785 -2.833167 -2.047343
31 6 0 -0.269938 -3.579173 -1.835978
32 1 0 -1.060050 -3.076365 -2.401544
33 1 0 0.084812 -4.449388 -2.387544
34 1 0 -0.711853 -3.893674 -0.886420
35 15 0 1.630170 2.928031 -0.713067
36 15 0 3.711460 -0.189918 -0.456603
37 6 0 2.659478 3.651623 0.641010
38 1 0 2.747356 4.736518 0.521934
39 1 0 2.187087 3.436533 1.603277
40 1 0 3.657850 3.213326 0.644903
41 6 0 2.469800 3.476366 -2.260614
42 1 0 1.839395 3.175352 -3.101799
43 1 0 2.607633 4.562530 -2.279198
44 1 0 3.442564 2.987435 -2.355986
45 6 0 0.155459 4.036032 -0.674728
46 1 0 -0.388353 3.911473 0.265752
47 1 0 0.473371 5.079897 -0.764859
48 1 0 -0.495986 3.773502 -1.511460
49 6 0 5.044779 0.854957 0.302937
50 1 0 5.995953 0.312301 0.326023
51 1 0 5.187014 1.771982 -0.275954
52 1 0 4.772993 1.126552 1.326684
53 6 0 3.892617 -1.754801 0.505894
54 1 0 4.944125 -2.055821 0.570467
55 1 0 3.486651 -1.627011 1.512130
56 1 0 3.323410 -2.533184 -0.004343
57 6 0 4.472966 -0.629820 -2.082604
58 1 0 5.486375 -1.025445 -1.954345
59 1 0 3.840542 -1.387360 -2.549793
60 1 0 4.512098 0.252258 -2.727945
61 6 0 -2.647912 -1.109894 -0.989268
62 6 0 -3.052515 -0.968889 -2.336620
63 6 0 -3.289773 -2.132061 -0.247159
64 6 0 -3.994426 -1.824368 -2.911588
65 1 0 -2.659043 -0.163676 -2.945518
66 6 0 -4.236597 -2.975354 -0.819641
67 1 0 -3.042735 -2.248548 0.803743
68 6 0 -4.590042 -2.841451 -2.165500
69 1 0 -4.276279 -1.675585 -3.950974
70 1 0 -4.702484 -3.743923 -0.208115
71 1 0 -5.325707 -3.501446 -2.615073
72 6 0 -0.666950 0.961001 -2.419304
73 8 0 -0.638664 2.014423 -3.064716
74 1 0 -0.463823 0.002119 -2.936897
75 6 0 -0.131906 -2.482218 2.079002
76 1 0 -0.860349 -2.764747 1.327077
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-in1II-rh
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 -1.986981 0.362521 1.322556
2 6 0 -3.203632 0.864526 1.823901
3 6 0 -4.159902 1.321193 0.944052
4 6 0 -3.905863 1.279252 -0.450106
5 6 0 -2.711825 0.797172 -0.923934
6 6 0 -0.469765 -0.202119 -0.292439
7 6 0 -0.021181 -0.476171 1.042679
8 1 0 -3.344456 0.872996 2.898487
9 1 0 -5.102482 1.713919 1.310924
10 1 0 -4.646856 1.627864 -1.160612
11 1 0 -2.456102 0.761860 -1.970594
12 6 0 1.254109 -1.022314 1.568071
13 6 0 1.876785 -0.322352 2.617301
14 6 0 2.953872 -2.745939 1.779332
15 6 0 3.033832 -0.812435 3.218503
16 1 0 1.433013 0.606306 2.961425
17 6 0 3.576996 -2.029721 2.800977
18 1 0 3.363281 -3.696881 1.451141
19 1 0 3.505253 -0.249480 4.019412
20 1 0 4.475096 -2.418843 3.272576
21 7 0 -0.935113 -0.126566 1.983477
22 7 0 -1.753753 0.350991 -0.053879
23 6 0 -0.951255 -1.943471 -1.617623
24 6 0 -0.896682 -1.273766 -2.881558
25 1 0 -0.041826 -2.470679 -1.343994
26 1 0 -1.807130 -0.803207 -3.247522
27 6 0 -2.176396 -2.439291 -0.977121
28 6 0 -2.081756 -3.145245 0.240857
29 6 0 -3.445383 -2.294770 -1.570516
30 6 0 -3.213512 -3.680547 0.843569
31 1 0 -1.111397 -3.261161 0.713791
32 6 0 -4.578686 -2.834686 -0.964244
33 1 0 -3.540204 -1.796989 -2.528965
34 6 0 -4.468055 -3.523651 0.243878
35 1 0 -3.120614 -4.221108 1.780554
36 1 0 -5.547892 -2.723971 -1.441543
37 1 0 -5.352141 -3.944809 0.713671
38 6 0 -0.076904 -1.792022 -3.991329
39 8 0 -0.316912 -1.517195 -5.163887
40 1 0 0.763155 -2.449209 -3.715698
41 6 0 1.805507 -2.245564 1.165337
42 1 0 1.344088 -2.813681 0.368879
43 6 0 0.704856 2.517855 -1.649674
44 6 0 2.000164 1.988664 -1.918815
45 6 0 2.030884 1.571342 -3.325836
46 6 0 0.757823 1.808885 -3.885843
47 6 0 -0.099243 2.342148 -2.838022
48 6 0 0.269186 3.189328 -0.381439
49 1 0 0.486860 4.264124 -0.427331
50 1 0 -0.803503 3.076553 -0.209679
51 1 0 0.791441 2.781314 0.486546
52 6 0 -1.442695 2.959991 -3.096635
53 1 0 -1.316206 3.941488 -3.573350
54 1 0 -2.046626 2.351455 -3.776486
55 1 0 -2.011497 3.115653 -2.178207
56 6 0 0.343605 1.589644 -5.310204
57 1 0 1.207125 1.632918 -5.978386
58 1 0 -0.130575 0.610060 -5.453093
59 1 0 -0.362766 2.362676 -5.629072
60 6 0 3.250572 1.067791 -4.031823
61 1 0 3.991663 1.871756 -4.127928
62 1 0 3.706058 0.245497 -3.473509
63 1 0 3.011377 0.700805 -5.031821
64 6 0 3.190553 1.990104 -1.009633
65 1 0 3.738316 1.048206 -1.092028
66 1 0 3.872789 2.810279 -1.270425
67 1 0 2.898333 2.115016 0.035252
68 45 0 0.535026 0.298274 -2.085258
69 17 0 2.320700 -1.396490 -1.843843
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-IIin2-rh
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 -3.368367 -0.861706 -1.617434
2 6 0 -4.347369 -1.726344 -2.161343
3 6 0 -4.594682 -2.935285 -1.559758
4 6 0 -3.874980 -3.303941 -0.388332
5 6 0 -2.918072 -2.478294 0.132297
6 6 0 -1.744048 -0.251003 -0.174838
7 6 0 -2.039187 0.761722 -1.107669
8 1 0 -4.885526 -1.388203 -3.039303
9 1 0 -5.346564 -3.605841 -1.962461
10 1 0 -4.081556 -4.240851 0.116144
11 1 0 -2.361558 -2.706462 1.029669
12 6 0 -1.486574 2.122249 -1.265028
13 6 0 -1.728132 2.804608 -2.474372
14 6 0 -0.255618 4.072871 -0.480881
15 6 0 -1.228293 4.085899 -2.686211
16 1 0 -2.314385 2.306459 -3.238056
17 6 0 -0.479425 4.725163 -1.693060
18 1 0 0.302770 4.564170 0.310788
19 1 0 -1.424460 4.589255 -3.629108
20 1 0 -0.089012 5.725504 -1.858174
21 7 0 -3.006537 0.356130 -1.989839
22 7 0 -2.636858 -1.281471 -0.491383
23 6 0 -0.793721 -0.379837 0.969127
24 6 0 -0.086885 -1.633167 1.060517
25 1 0 1.359591 1.336829 0.677888
26 1 0 -0.248648 -2.333877 0.246885
27 6 0 -1.179014 0.399528 2.205408
28 6 0 -0.345536 0.551892 3.331026
29 6 0 -2.472827 0.953489 2.291096
30 6 0 -0.785047 1.209302 4.478319
31 1 0 0.672373 0.191347 3.306557
32 6 0 -2.913513 1.610359 3.439208
33 1 0 -3.148077 0.865215 1.447992
34 6 0 -2.073133 1.741353 4.543739
35 1 0 -0.107676 1.312173 5.321256
36 1 0 -3.919452 2.019659 3.464360
37 1 0 -2.412508 2.255324 5.438291
38 6 0 0.439061 -2.347623 2.231342
39 8 0 0.819391 -3.513866 2.152031
40 1 0 0.492166 -1.817571 3.196410
41 6 0 -0.758396 2.787855 -0.264642
42 1 0 -0.610639 2.324257 0.700392
43 6 0 2.967845 0.338915 -1.387979
44 6 0 3.537671 -0.807900 -0.655534
45 6 0 2.734474 -1.937097 -0.885580
46 6 0 1.582907 -1.473746 -1.661534
47 6 0 1.817134 -0.097352 -2.073424
48 6 0 3.630539 1.678514 -1.501700
49 1 0 2.936912 2.436345 -1.872586
50 1 0 4.010987 2.018528 -0.535162
51 1 0 4.480150 1.630132 -2.195669
52 6 0 1.031739 0.645174 -3.108773
53 1 0 1.431220 0.431128 -4.110007
54 1 0 -0.019746 0.351995 -3.107035
55 1 0 1.071083 1.724947 -2.956506
56 6 0 0.554693 -2.379431 -2.271652
57 1 0 0.892382 -2.702622 -3.265429
58 1 0 0.400407 -3.280023 -1.672842
59 1 0 -0.406935 -1.877609 -2.401045
60 6 0 2.956625 -3.346884 -0.422903
61 1 0 2.984249 -4.033514 -1.278204
62 1 0 3.909470 -3.441491 0.103004
63 1 0 2.171283 -3.690192 0.260844
64 6 0 4.805868 -0.735668 0.138866
65 1 0 4.968389 -1.642994 0.723953
66 1 0 5.670016 -0.599736 -0.524221
67 1 0 4.786976 0.112569 0.829608
68 45 0 1.403786 -0.335692 0.219269
69 17 0 2.300009 1.652818 1.816894
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-IVin3
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 2.463415 -2.150365 -1.230532
2 6 0 2.117538 -2.841107 -2.408045
3 6 0 0.959283 -2.493505 -3.058170
4 6 0 0.151895 -1.438541 -2.557600
5 6 0 0.480625 -0.726397 -1.419857
6 6 0 2.225548 -0.638758 0.416651
7 6 0 3.365439 -1.451332 0.590782
8 1 0 2.775712 -3.632507 -2.746334
9 1 0 0.652530 -3.020742 -3.956984
10 1 0 -0.777943 -1.204894 -3.061546
11 6 0 4.314446 -1.515819 1.719673
12 6 0 5.665390 -1.801596 1.462034
13 6 0 4.808256 -1.511388 4.098363
14 6 0 6.577528 -1.921599 2.508026
15 1 0 5.982437 -1.935161 0.433019
16 6 0 6.153231 -1.772588 3.830869
17 1 0 4.465636 -1.414391 5.124775
18 1 0 7.620587 -2.136139 2.291626
19 1 0 6.863770 -1.869477 4.647119
20 7 0 3.505467 -2.342863 -0.424562
21 7 0 1.628007 -1.115240 -0.759348
22 6 0 1.687661 0.461824 1.196331
23 6 0 0.326362 0.668706 1.324537
24 1 0 -0.402987 -0.548488 1.509455
25 46 0 -0.886053 0.584777 -0.666861
26 6 0 -1.714239 -2.060051 0.967955
27 8 0 -0.849516 -1.626345 1.818751
28 8 0 -2.126244 -1.423245 -0.028944
29 6 0 -2.237259 -3.459984 1.235835
30 1 0 -2.550342 -3.549995 2.279556
31 1 0 -3.066988 -3.693460 0.567378
32 1 0 -1.428215 -4.180004 1.075210
33 15 0 -2.948086 1.669643 0.118191
34 15 0 -0.505343 1.921847 -2.559964
35 6 0 -3.064867 3.428405 0.674795
36 1 0 -4.104477 3.685238 0.903085
37 1 0 -2.461935 3.528580 1.580640
38 1 0 -2.690394 4.113390 -0.088254
39 6 0 -4.243093 1.550712 -1.198850
40 1 0 -4.305096 0.510880 -1.530529
41 1 0 -5.221472 1.866708 -0.822416
42 1 0 -3.980964 2.172506 -2.059407
43 6 0 -3.740048 0.774035 1.517104
44 1 0 -3.048259 0.792489 2.361257
45 1 0 -4.681414 1.261137 1.792002
46 1 0 -3.925854 -0.255874 1.211609
47 6 0 -1.008046 3.694963 -2.404488
48 1 0 -0.655168 4.270814 -3.266288
49 1 0 -2.095643 3.781015 -2.348143
50 1 0 -0.577900 4.117339 -1.493107
51 6 0 1.291082 2.091722 -2.944990
52 1 0 1.452638 2.780956 -3.780329
53 1 0 1.811464 2.465181 -2.059222
54 1 0 1.700689 1.111081 -3.194778
55 6 0 -1.223848 1.477662 -4.207411
56 1 0 -0.943897 2.213640 -4.968677
57 1 0 -0.856118 0.496189 -4.512426
58 1 0 -2.313510 1.431012 -4.134324
59 6 0 2.688036 1.352475 1.858829
60 6 0 2.514629 1.807046 3.178117
61 6 0 3.812549 1.804563 1.143441
62 6 0 3.425288 2.690816 3.756795
63 1 0 1.675485 1.441734 3.761314
64 6 0 4.715006 2.696503 1.717723
65 1 0 3.970486 1.452153 0.128978
66 6 0 4.526336 3.142323 3.028446
67 1 0 3.277006 3.019034 4.781639
68 1 0 5.570655 3.040744 1.143422
69 1 0 5.236401 3.829605 3.479536
70 6 0 -0.138948 1.891272 1.979410
71 8 0 -1.116673 1.994046 2.724108
72 1 0 0.431242 2.814936 1.724082
73 6 0 3.894402 -1.387752 3.052594
74 1 0 2.845361 -1.216495 3.268843
---------------------------------------------------------------------
Optimized Cartesian coordinates for ts-Vin4
---------------------------------------------------------------------
Center Atomic Atomic Coordinates (Angstroms)
Number Number Type X Y Z
---------------------------------------------------------------------
1 6 0 0.578107 -2.806538 0.473005
2 6 0 -0.450754 -3.697350 0.792405
3 6 0 -1.732589 -3.167508 0.963159
4 6 0 -2.025393 -1.808033 0.735975
5 6 0 -1.035736 -0.926881 0.271964
6 6 0 1.370230 -0.710907 0.284156
7 6 0 2.423032 -1.679831 0.314402
8 1 0 -0.225367 -4.742040 0.968793
9 1 0 -2.540511 -3.831291 1.259640
10 1 0 -3.042992 -1.449739 0.844491
11 6 0 3.873065 -1.474836 0.190512
12 6 0 4.754471 -2.482738 0.619954
13 6 0 5.784505 -0.157330 -0.527781
14 6 0 6.130940 -2.323095 0.484394
15 1 0 4.336344 -3.384533 1.053640
16 6 0 6.652197 -1.159036 -0.087403
17 1 0 6.180868 0.745966 -0.982766
18 1 0 6.800139 -3.108465 0.824837
19 1 0 7.726450 -1.036092 -0.193406
20 7 0 1.923514 -2.932946 0.438048
21 7 0 0.224114 -1.484993 0.262983
22 6 0 1.060436 0.631139 0.662732
23 6 0 -0.308129 0.966322 0.602868
24 46 0 -1.474116 0.561526 -1.093554
25 15 0 -2.460040 -0.880876 -2.797217
26 15 0 -1.966302 2.826451 -1.892129
27 6 0 -1.442848 -2.408256 -3.049216
28 1 0 -1.912023 -3.093092 -3.763957
29 1 0 -0.452297 -2.128925 -3.418639
30 1 0 -1.313941 -2.919370 -2.092032
31 6 0 -4.115670 -1.605358 -2.385670
32 1 0 -4.865460 -0.810093 -2.343411
33 1 0 -4.426992 -2.352591 -3.123402
34 1 0 -4.062712 -2.076307 -1.401217
35 6 0 -2.738136 -0.358064 -4.557133
36 1 0 -1.809622 0.041229 -4.974235
37 1 0 -3.072801 -1.197218 -5.176498
38 1 0 -3.496798 0.428575 -4.596944
39 6 0 -3.673883 3.318502 -1.377538
40 1 0 -3.911980 4.344715 -1.677713
41 1 0 -4.404746 2.636853 -1.822187
42 1 0 -3.728842 3.223821 -0.290736
43 6 0 -0.968114 4.241605 -1.236232
44 1 0 -1.364210 5.202613 -1.581794
45 1 0 -0.992661 4.214987 -0.145449
46 1 0 0.068906 4.146598 -1.570929
47 6 0 -1.972614 3.246193 -3.701367
48 1 0 -2.261546 4.288836 -3.873317
49 1 0 -0.974541 3.082673 -4.118502
50 1 0 -2.671231 2.596511 -4.233553
51 6 0 2.065148 1.528830 1.287445
52 6 0 2.941690 1.062600 2.284755
53 6 0 2.140024 2.882453 0.914940
54 6 0 3.857525 1.922171 2.886470
55 1 0 2.889247 0.024817 2.596205
56 6 0 3.061235 3.741435 1.514605
57 1 0 1.476573 3.251053 0.140198
58 6 0 3.923346 3.264269 2.502595
59 1 0 4.519225 1.544277 3.660581
60 1 0 3.105350 4.782893 1.208257
61 1 0 4.640449 3.931803 2.971818
62 6 0 -0.889466 1.926485 1.531101
63 8 0 -2.039992 2.363442 1.481312
64 1 0 -0.225883 2.246886 2.361735
65 6 0 4.406309 -0.313579 -0.393361
66 1 0 3.737138 0.461914 -0.750604
---------------------------------------------------------------------

Table S1: Calculated relative energies (all in kcal mol-1, relative to isolated species) for the ZPE-corrected Gibbs free energies (ΔGgas), Gibbs free energies for all species in solution phase (ΔGsol) at 393 K, 413 K by B3LYP/6-311++G(d,p)//B3LYP/6-31G(d) method and difference between absolute energy.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.